 PDF(15248 KB)
PDF(15248 KB)


Application of Advanced Artificial Intelligence Technology in New Drug Discovery
Zhonghua Wang, Yichu Wu, Zhongshan Wu, Ranran Zhu, Yang Yang, Fanhong Wu
Prog Chem ›› 2023, Vol. 35 ›› Issue (10) : 1505-1518.
 PDF(15248 KB)
PDF(15248 KB)
 PDF(15248 KB)
PDF(15248 KB)
Application of Advanced Artificial Intelligence Technology in New Drug Discovery
In recent years, the discovery of new drugs driven by advanced artificial intelligence (AI) has attracted much attention. Advanced artificial intelligence algorithms (machine learning and deep learning) have been gradually applied in various scenarios of new drug discovery, such as representation learning task (molecular descriptor), prediction task (drug target binding affinity prediction, crystal structure prediction and molecular basic properties prediction) and generation task (molecular conformation generation and drug molecular generation). This technology can significantly reduce the cost and time of new drug development, improve the efficiency of drug development, and reduce the costs and risks associated with preclinical and clinical trials. This review summarizes the application of advanced artificial intelligence technology in new drug discovery in recent years, to help understand the research progress and future development trend in this field, and to facilitate the discovery of innovative drugs.
1 Introduction
2 Artificial intelligence
2.1 Convolutional neural network
2.2 Recurrent neural network
2.3 Graph neural network
2.4 Generative adversarial network
2.5 Variational auto encoder
2.6 Diffusion model
2.7 Transformer model
3 The application of artificial intelligence in drug discovery
3.1 Data resources and open-source tools
3.2 Artificial intelligence technology drives molecular representation learning tasks
3.3 Artificial intelligence technology drives predictive tasks
3.4 Artificial intelligence technology drives generation tasks
4 Conclusion and outlook
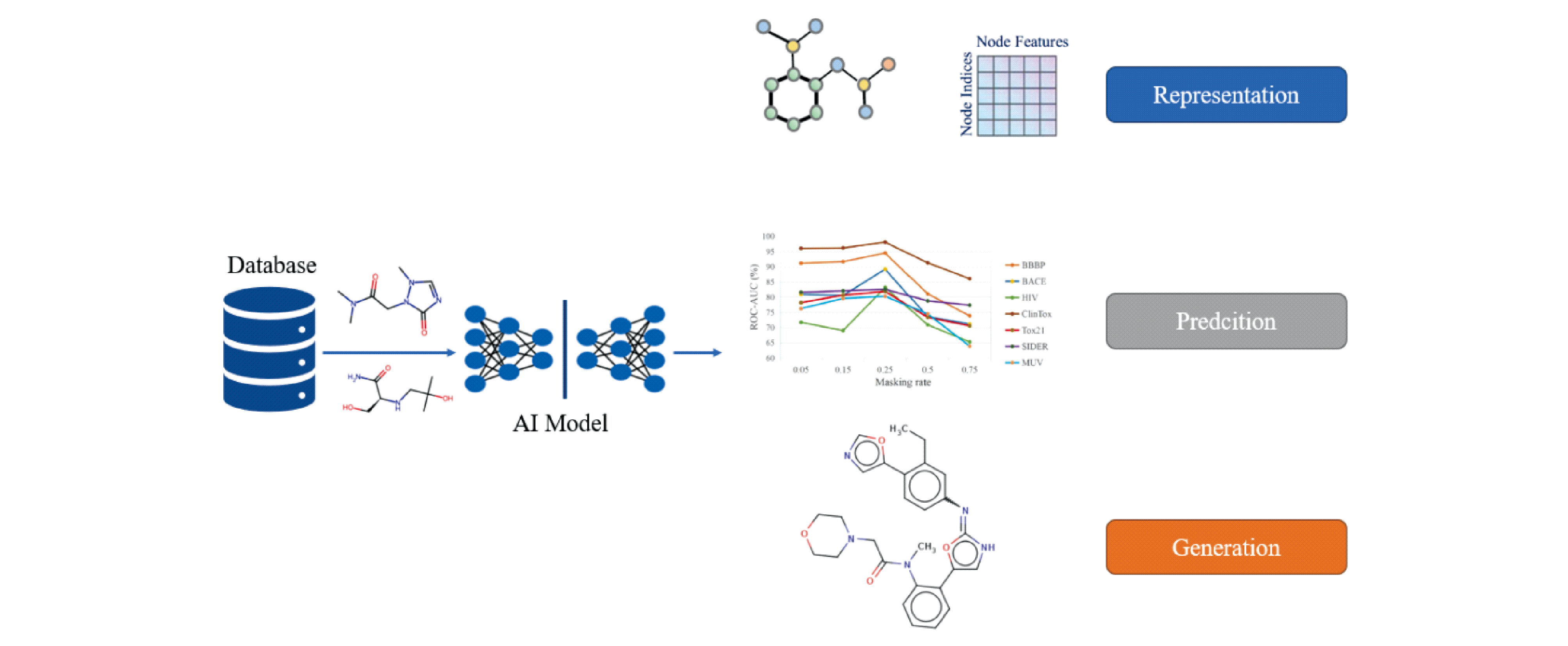
artificial intelligence / new drug discovery / deep learning / representation learning / task application
| [1] |
|
| [2] |
|
| [3] |
|
| [4] |
|
| [5] |
|
| [6] |
|
| [7] |
|
| [8] |
|
| [9] |
|
| [10] |
|
| [11] |
|
| [12] |
|
| [13] |
|
| [14] |
|
| [15] |
|
| [16] |
|
| [17] |
|
| [18] |
|
| [19] |
|
| [20] |
|
| [21] |
|
| [22] |
|
| [23] |
|
| [24] |
|
| [25] |
|
| [26] |
|
| [27] |
|
| [28] |
|
| [29] |
|
| [30] |
|
| [31] |
|
| [32] |
|
| [33] |
|
| [34] |
|
| [35] |
|
| [36] |
|
| [37] |
|
| [38] |
|
| [39] |
|
| [40] |
|
| [41] |
|
| [42] |
|
| [43] |
|
| [44] |
OpenAI. arXiv preprint arXiv: 2303.08774, 2023.
|
| [45] |
|
| [46] |
|
| [47] |
|
| [48] |
|
| [49] |
|
| [50] |
|
| [51] |
|
| [52] |
|
| [53] |
|
| [54] |
|
| [55] |
|
| [56] |
|
| [57] |
|
| [58] |
|
| [59] |
|
| [60] |
|
| [61] |
|
| [62] |
|
| [63] |
|
| [64] |
|
| [65] |
|
| [66] |
|
| [67] |
|
| [68] |
|
| [69] |
|
| [70] |
|
| [71] |
|
| [72] |
|
| [73] |
|
| [74] |
|
| [75] |
|
| [76] |
|
| [77] |
|
| [78] |
|
| [79] |
|
| [80] |
|
| [81] |
|
| [82] |
|
| [83] |
|
| [84] |
|
| [85] |
|
| [86] |
|
| [87] |
|
| [88] |
|
| [89] |
|
| [90] |
|
| [91] |
|
| [92] |
|
| [93] |
|
| [94] |
|
| [95] |
|
| [96] |
|
| [97] |
|
| [98] |
|
| [99] |
|
| [100] |
|
| [101] |
|
| [102] |
|
| [103] |
|
| [104] |
|
| [105] |
|
| [106] |
|
| [107] |
|
| [108] |
|
| [109] |
|
| [110] |
|
/
| 〈 |
|
〉 |