 PDF(1033 KB)
PDF(1033 KB)


Research Methods for Liquid-Liquid Phase Separation of Biological Macromolecules
Chubin Zhao, Hailin Wang
Prog Chem ›› 2023, Vol. 35 ›› Issue (10) : 1486-1491.
 PDF(1033 KB)
PDF(1033 KB)
 PDF(1033 KB)
PDF(1033 KB)
Research Methods for Liquid-Liquid Phase Separation of Biological Macromolecules
The liquid-liquid phase separation of biological macromolecules is widely observed in various biological systems, and has become an emerging research focus of life science in recent years. Biological macromolecules are continuously enriched through multivalent interaction. When the molecular concentration reaches the dissolution threshold in solution, they will be precipitated from solution in the form of liquid-liquid phase separation. It is closely related to many important biological processes in cells (such as the formation of membraneless organelles). With the deepening of research on phase separation, its research methods are also developing and improving. Based on the principle and characteristics of phase separation, this paper introduces some commonly used research methods of phase separation. It provides the method basis for the subsequent phase separation research and promotes the further development of phase separation techniques and methods.
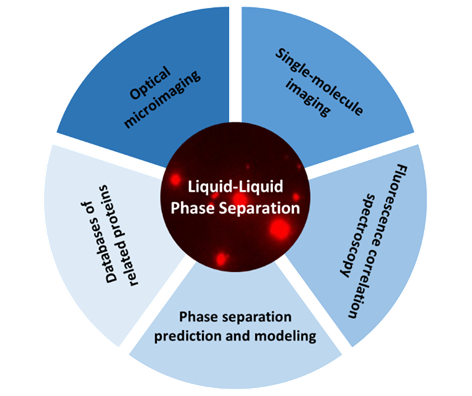
1 Principle and characteristics of liquid-liquid separation
2 Imaging technique for liquid-liquid phase separation
2.1 Optical microimaging
2.2 Single-molecule fluorescence imaging
2.3 Fluorescence correlation spectroscopy
3 Theoretical prediction for liquid-liquid separation
3.1 Phase separation prediction and modeling
3.2 Databases of phase separation related proteins
4 Conclusion and outlook
liquid-liquid phase separation / optical microscopy / fluorescence correlation spectroscopy / molecular simulation
| [1] |
|
| [2] |
(郜一飞, 李丕龙. 中国细胞生物学学报, 2019, 41(2): 185.).
|
| [3] |
|
| [4] |
|
| [5] |
|
| [6] |
|
| [7] |
|
| [8] |
|
| [9] |
|
| [10] |
|
| [11] |
|
| [12] |
|
| [13] |
|
| [14] |
|
| [15] |
|
| [16] |
|
| [17] |
|
| [18] |
|
| [19] |
|
| [20] |
|
| [21] |
|
| [22] |
(张祥翔. 光学仪器, 2015, 37(6): 550.).
|
| [23] |
(关苑君, 马显才. 中山大学学报(医学科学版), 2022, 43(3): 504.).
|
| [24] |
|
| [25] |
|
| [26] |
|
| [27] |
|
| [28] |
|
| [29] |
|
| [30] |
|
| [31] |
|
| [32] |
|
| [33] |
|
| [34] |
|
| [35] |
|
| [36] |
|
| [37] |
|
| [38] |
|
| [39] |
|
| [40] |
|
| [41] |
|
| [42] |
|
| [43] |
|
| [44] |
|
| [45] |
(张鹏程, 方文玉, 鲍磊, 康文斌. 物理学报, 2020, 69(13): 278.).
|
| [46] |
|
| [47] |
|
| [48] |
|
| [49] |
|
| [50] |
|
| [51] |
|
| [52] |
|
| [53] |
|
| [54] |
|
| [55] |
|
| [56] |
|
| [57] |
|
| [58] |
|
/
| 〈 |
|
〉 |