 PDF(420770 KB)
PDF(420770 KB)


Optimizing Metabolic Pathways by Using Bioretrosynthesis Tools
Liu Fufeng, Liu Xuzhi, Li Jinbi, Lu Fuping
Prog Chem ›› 2024, Vol. 36 ›› Issue (4) : 501-510.
 PDF(420770 KB)
PDF(420770 KB)
 PDF(420770 KB)
PDF(420770 KB)
Optimizing Metabolic Pathways by Using Bioretrosynthesis Tools
Biocatalysis has become an important technology In the field of biosynthesis because of its mild reaction conditions,high efficiency,high specificity and low price.There are a series of highly integrated metabolic networks in the biosynthesis system,and the study of multi-enzyme catalytic system has become an inevitable trend in the field of biosynthesis,so it is of great significance to explore the unknown multi-enzyme synthesis path based on the known products.in this review,the concepts of multi-enzyme system and retrosynthesis process are introduced.and the design methods,advantages and disadvantages of retrosynthesis tools are summarized.Then the tools are divided into host-based and host-less tools.For each of these two types,some representative retrosynthesis tools are listed to analyze their respective design processes and differences.Finally,the possibility of artificial intelligence-assisted multi-enzyme system is discussed and the optimization and development of multi-enzyme pathway construction tools are forecasted。
1 Introduction
2 Multienzyme catalysis
3 Methods for building retrosynthesis tools
4 Introduction to the retrosynthesis tools
4.1 Host-based retrosynthetic tools
4.2 Host-free retrosynthetic tools
5 Artificial intelligence fuels the development of multi-enzyme systems
6 Conclusion and outlook
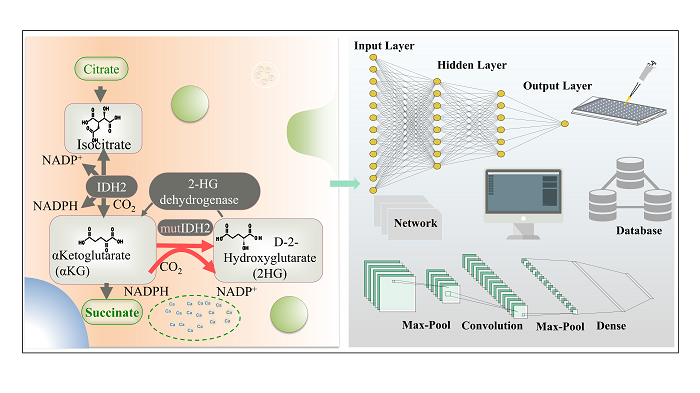
multi-enzyme catalysis / path design / retrosynthesis / biological retrosynthesis tool
| [1] |
|
| [2] |
( 王慧悦, 胡欣, 胡玉静, 朱宁, 郭凯. 化学进展, 2022, 34(8): 1796.)
|
| [3] |
( 李红, 史晓丹, 李洁龄. 化学进展, 2022, 34(3): 568.)
|
| [4] |
( 武江洁星, 魏辉. 化学进展, 2021, 33(1): 42.)
|
| [5] |
|
| [6] |
|
| [7] |
|
| [8] |
( 赵自通, 张真真, 梁志宏. 化学进展, 2022, 34(11): 2386.)
|
| [9] |
( 黄文倩, 王迎夏, 田维圣, 王娟, 屠鹏飞, 王晓晖, 史社坡, 刘晓. 中国中药杂志, 2023, 48(2): 336.)
|
| [10] |
|
| [11] |
|
| [12] |
|
| [13] |
|
| [14] |
( 曾涛, 巫瑞波. 合成生物学, 2023, 4(3): 535.)
|
| [15] |
|
| [16] |
|
| [17] |
|
| [18] |
|
| [19] |
|
| [20] |
|
| [21] |
|
| [22] |
|
| [23] |
|
| [24] |
|
| [25] |
|
| [26] |
|
| [27] |
( 魏奕新, 韩一蕾, 卢滇楠, 邱彤. 清华大学学报(自然科学版), 2023, 63(5): 697.)
|
| [28] |
|
| [29] |
|
| [30] |
|
| [31] |
|
| [32] |
( 李思徵. 兰州大学硕士论文, 2023.)
|
| [33] |
( 陈颖莹, 荣丹琪, 李元晶, 赵鸿萍. 化学通报, 2022, 85(08): 951.)
|
| [34] |
|
| [35] |
|
| [36] |
|
| [37] |
|
| [38] |
|
| [39] |
|
| [40] |
|
| [41] |
|
| [42] |
|
| [43] |
|
| [44] |
|
| [45] |
|
| [46] |
|
| [47] |
|
| [48] |
|
| [49] |
( 丁德武, 丁彦蕊, 蔡宇杰, 陈守文, 须文波. 计算机与应用化学, 2008, 25(1): 4.)
|
| [50] |
|
| [51] |
|
| [52] |
|
| [53] |
|
| [54] |
|
| [55] |
|
| [56] |
|
| [57] |
|
| [58] |
|
| [59] |
|
| [60] |
|
| [61] |
|
| [62] |
|
| [63] |
|
| [64] |
|
| [65] |
|
| [66] |
|
| [67] |
|
| [68] |
|
| [69] |
|
| [70] |
|
| [71] |
|
| [72] |
|
| [73] |
|
| [74] |
|
| [75] |
|
| [76] |
|
| [77] |
|
| [78] |
|
| [79] |
( 丁邵珍, 江小琴, 孟超, 孙丽霞, 王正权, 杨弘宾, 沈国文, 夏宁. 中国科学: 化学, 2023, 53(01): 66.)
|
| [80] |
|
| [81] |
|
| [82] |
|
| [83] |
|
| [84] |
|
| [85] |
|
| [86] |
|
| [87] |
|
| [88] |
|
/
| 〈 |
|
〉 |