 PDF(3841 KB)
PDF(3841 KB)


Intracellular Single Strand DNA and High-Throughput Analysis Techniques
Ruiqi Li, Weiyi Lai, Hailin Wang
Prog Chem ›› 2024, Vol. 36 ›› Issue (9) : 1283-1290.
 PDF(3841 KB)
PDF(3841 KB)
 PDF(3841 KB)
PDF(3841 KB)
Intracellular Single Strand DNA and High-Throughput Analysis Techniques
During many life processes such as replication,transcription,double-strand breaks repair and so on,double-stranded DNA will temporarily unwind and form single strand DNA(ssDNA).ssDNA may affect genomic stability and may also participate in the formation of non-B DNA structure,which in turn regulates and influences many key cellular processes.This review briefly describes the causes of the formation of single-stranded DNA,the structures containing single-stranded DNA and their possible functions in cells,and summarizes some high-throughput analysis techniques of single-stranded DNA,which provides the method inspiration for the subsequent ssDNA research and promotes the further development of ssDNA analysis techniques and methods。
1 Overview of ssDNA
2 Formation and function of ssDNA
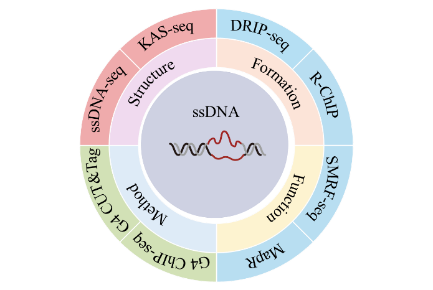
3 ssDNA sequencing methods
3.1 ssDNA-seq
3.2 KAS-seq
3.3 DRIP-seq
3.4 R-ChIP
3.5 SMRF-seq
3.6 MapR
3.7 G4 ChIP-seq
3.8 G4 CUT&Tag
4 Conclusion and outlook
single strand DNA / R-loop / G-quadruplex / high-throughput sequencing
| [1] |
|
| [2] |
|
| [3] |
|
| [4] |
|
| [5] |
|
| [6] |
|
| [7] |
|
| [8] |
|
| [9] |
|
| [10] |
|
| [11] |
|
| [12] |
|
| [13] |
|
| [14] |
|
| [15] |
|
| [16] |
|
| [17] |
|
| [18] |
|
| [19] |
|
| [20] |
|
| [21] |
|
| [22] |
|
| [23] |
|
| [24] |
|
| [25] |
|
| [26] |
|
| [27] |
|
| [28] |
|
| [29] |
|
| [30] |
|
| [31] |
|
| [32] |
|
| [33] |
|
| [34] |
|
| [35] |
|
| [36] |
|
| [37] |
|
| [38] |
|
| [39] |
|
| [40] |
|
| [41] |
|
| [42] |
|
| [43] |
|
| [44] |
|
| [45] |
|
| [46] |
|
| [47] |
|
| [48] |
|
| [49] |
|
| [50] |
|
| [51] |
|
| [52] |
|
| [53] |
|
| [54] |
|
| [55] |
|
| [56] |
|
| [57] |
|
| [58] |
|
| [59] |
|
| [60] |
|
| [61] |
|
| [62] |
|
| [63] |
|
| [64] |
|
| [65] |
|
| [66] |
|
| [67] |
|
| [68] |
|
| [69] |
|
| [70] |
|
| [71] |
|
| [72] |
|
| [73] |
|
| [74] |
|
| [75] |
|
| [76] |
|
/
| 〈 |
|
〉 |